Exercise Four: Test Different Meshes
In this exercise, we will explore how to modify the mesh resolution in NextGenPB and observe its impact on the calculated electrostatic potential.
Step 1 – Prepare the Inputs
Navigate to the working directory for this exercise:
cd ~/ngpb_tutorial/ex4
Copy the necessary input files:
cp ../NextGenPB/data/1CCM.pdb .
cp ../NextGenPB/data/charge.crg .
cp ../NextGenPB/data/radius.siz .
Create Parameter Files
We will prepare multiple parameter files to test different mesh resolutions by varying the scale, the perfil, and by applying random displacement of the mesh center.
A full description of mesh options is available in the Guide
Fine Mesh (fine_mesh.prm)
Create a file named fine_mesh.prm in the current directory with the following content:
[input]
filetype = pdb
filename = 1CCM.pdb
radius_file = radius.siz
charge_file = charge.crg
write_pqr = 1
name_pqr = 1CCM_out.pqr
[../]
[mesh]
mesh_shape = 0
perfil1 = 0.95
perfil2 = 0.2
scale = 4.0
[../]
[model]
bc_type = 1
molecular_dielectric_constant = 2 # Dielectric constant inside the molecule
solvent_dielectric_constant = 80 # Dielectric constant of the solvent (e.g., water)
ionic_strength = 0.145 # Ionic strength (mol/L)
T = 298.15 # Temperature in Kelvin
calc_energy = 2
calc_coulombic = 1
map_type = vtu # Output format: 'vtu' (for ParaView, vtk binary), 'oct' (Octbin internal format)
potential_map = 1 # 1 = write full potential map to file
[../]
This will produce a mesh with a grid scale in the region of perfil1 equal to 0.25 Å.
Random center (rand_center.prm)
Create a file named rand_center.prm in the current directory with the following content:
[input]
filetype = pdb
filename = 1CCM.pdb
radius_file = radius.siz
charge_file = charge.crg
write_pqr = 1
name_pqr = 1CCM_out.pqr
[../]
[mesh]
mesh_shape = 0
perfil1 = 0.95
perfil2 = 0.2
scale = 2.0
rand_center = 1
[../]
[model]
bc_type = 1
molecular_dielectric_constant = 2 # Dielectric constant inside the molecule
solvent_dielectric_constant = 80 # Dielectric constant of the solvent (e.g., water)
ionic_strength = 0.145 # Ionic strength (mol/L)
T = 298.15 # Temperature in Kelvin
calc_energy = 2
calc_coulombic = 1
map_type = vtu # Output format: 'vtu' (for ParaView, vtk binary), 'oct' (Octbin internal format)
potential_map = 1 # 1 = write full potential map to file
[../]
This will randomly shift the mesh center within an interval of one grid scale.
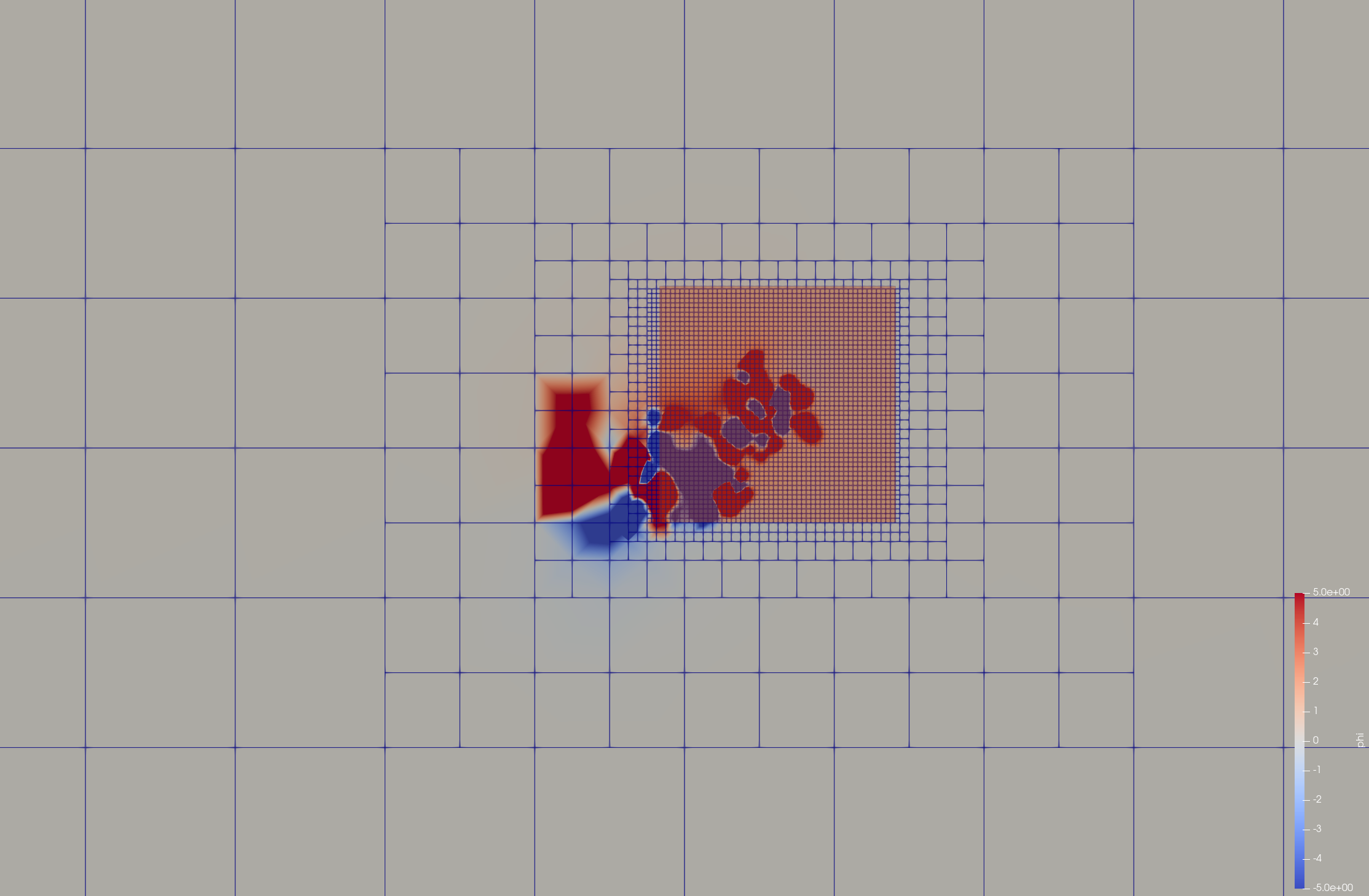
Focusing (focus.prm)
Create a file named focus.prm in the current directory with the following content:
[input]
filetype = pdb
filename = 1CCM.pdb
radius_file = radius.siz
charge_file = charge.crg
write_pqr = 1
name_pqr = 1CCM_out.pqr
[../]
[mesh]
mesh_shape = 3
perfil1 = 0.9
perfil2 = 0.2
scale = 2.0
cx_foc = 1 # X-center of focusing region
cy_foc = 10 # Y-center of focusing region
cz_foc = 5 # Z-center of focusing region
n_grid = 50 # Number of 1/scale-width intervals in focused zone
[../]
[model]
bc_type = 1
molecular_dielectric_constant = 2 # Dielectric constant inside the molecule
solvent_dielectric_constant = 80 # Dielectric constant of the solvent (e.g., water)
ionic_strength = 0.145 # Ionic strength (mol/L)
T = 298.15 # Temperature in Kelvin
calc_energy = 2
calc_coulombic = 1
map_type = vtu # Output format: 'vtu' (for ParaView, vtk binary), 'oct' (Octbin internal format)
potential_map = 1 # 1 = write full potential map to file
[../]
Step 2 – Run the Solver
Now launch the simulation using Apptainer:
Refined grid
apptainer exec --pwd /App --bind .:/App ../NextGenPB.sif mpirun -np 4 ngpb --prmfile fine_mesh.prm
Random center
apptainer exec --pwd /App --bind .:/App ../NextGenPB.sif mpirun -np 4 ngpb --prmfile rand_center.prm
Focusing
apptainer exec --pwd /App --bind .:/App ../NextGenPB.sif mpirun -np 4 ngpb --prmfile focus.prm
➡️ For more information, see the Guide.